Ideonella sakaiensis, a new species of the genus Ideonella, is a gram-negative, aerobic, rod-shaped bacterium. It is also known as plastic-eating bacteria.
- Plastic-eating bacteria can use plastic as their primary source of carbon and energy for growth.
- The age of plastics started with Leo Baekeland’s discovery of Bakelite in 1907. Since a wide range of synthetic polymers has been created, they are now commonplace and indispensable in human life and, regrettably, the environment.
- In landfills and other locations, there was 5,000 Mt of plastic waste in 2015. By 2050, that number may rise to 12,000 Mt.
- Ester bonds, which connect poly (ethylene terephthalate) monomers, can be hydrolyzed by a variety of hydrolytic enzymes found in nature. PET is theoretically more susceptible to natural degradation than polyolefin since there are no known enzymes that can directly break their C-C bonds.
- A PET-hydrolyzing enzyme was discovered due to the genome sequencing of I. sakaiensis.
- An amorphous PET film’s surface develops crater-like pitting, and the hydrolysis products are released into the aqueous environment.
- Since the I. sakaiensis enzyme has the highest catalytic preference for PET at room temperature compared to other known PHEs, it was named PET hydrolase (PETase).
- This bacterium uses PET as a major source of carbon and energy, breaking it down into CO2 and water.
Ideonella sakaiensis Discovery
- Among 250 samples from a PET-contaminated environment, including sediment, soil, wastewater, and activated sludge close to a plastic bottle recycling site, a team of researchers led by Kohei Oda of Kyoto Institute of Technology and Kenji Miyamoto of Keio University reported that they had isolated a novel bacterium, Ideonella sakaiensis 201-F6, from the environment.
- They looked for microorganisms that could grow primarily on low-crystallinity (1.9 percent) PET film using these samples.
- A distinct microbial consortium that developed on the PET film after being cultured in one sediment sample caused the PET film to change morphologically.
- The species Ideonella sakaiensis is a member of the Ideonella genus.
- The bacterium was discovered in Sakai, a city in Japan, hence the name of the organism.
- The bacteria were initially isolated from a group of microbes in the sediment sample, which also included protozoa and cells that resembled yeast.
- Cells appeared connected by appendages on the film and were also seen in the culture fluid.
- Between the cells and the film, there were shorter appendages that may have helped deliver secreted enzymes to the film.
- After six weeks at 30°C, the PET film had suffered significant damage and was almost completely degraded.
Ideonella sakaiensis Classification
- The concept of plastic degradation by Ideonella sakaiensis, which belongs to the genus Ideonella and the family Comamonadaceae, was developed in response to the search for a biological system that could break down plastic debris.
- Members of the Comamonadaceae family are found together in a phylogenetic cluster in the Betaproteobacteria 16S rRNA phylogeny.
Domain | Bacteria |
Phylum | Pseudomonadota |
Class | Betaproteobacteria |
Order | Burkholderiales |
Family | Comamonadaceae |
Genus | Ideonella |
Species | Ideonella sakaiensis |
Habitat of Ideonella sakaiensis
- Ideonella sakaiensis exist in a variety of natural habitats as well as those created by humans, and they are indifferent to whether an area is clean or polluted.
- Some examples of such habitats include soil, fresh and groundwater, activated sludge, and industrial processing water.
- Due to the wide variety of habitats, the morphological and physiological characteristics of the individual features of the genera vary greatly within the species themselves.
Morphology of Ideonella sakaiensis
- Ideonella sakaiensis is gram-negative, aerobic, and rod-shaped.
- They are about 0.6 to 0.8 μm in width and 1.2-1.5 μm in length.
- The polar flagellum on the cells allows for motility.
- It can grow between temperatures of 15-42°C (28°C best) and the pH range of 5.5–9.0 (pH 7–7.5 is ideal).
- Genomic DNA had a G+C content of 70.4 mol%.
- For this bacterium, 3% (w/v) NaCl is lethal.
- Ideonella sakaiensis can adhere to PET surfaces and grow, using its thin appendages to hold onto the PET film.
- These extensions may deliver enzymes that break down PET to the PET film.
Cultural characteristics of Ideonella sakaiensis
Ideonella sakaiensis is an excellent choice for growth in environments with high concentrations of accumulated plastics because, interestingly, it uses PET as its sole source of carbon and energy for growth.
NBRC no.802 agar
- After two days of incubation at 30°C, colonies were 0.5-1.0 mm in diameter, circular, raised, translucent, with an entire margin, and non-pigmented or cream.
Biochemical characteristics of Ideonella sakaiensis
The biochemical characteristics of Ideonella sakaiensis can be tabulated as follows;
S.N. | Basic characteristics | Ideonella sakaiensis |
1. | Gram staining | Negative (-) |
2. | Shape | Rods |
3. | Spore | Negative (-) |
4. | Motility | Motile |
5. | Nitrate reduction | Negative (-) |
6. | Catalase | Positive (+) |
7. | Oxidase | Positive (+) |
8. | Indole formation | Negative (-) |
9. | Pigmentation | Negative (-) |
Fermentation of
S.N. | Sugars | Ideonella sakaiensis |
1. | Glucose | Negative (-) |
2. | Maltose | Positive (+) |
Enzymatic reactions
S.N. | Enzymes | Ideonella sakaiensis |
1. | Galactosidase | Negative (-) |
Mode of action of Ideonella sakaiensis
1) PETase enzyme
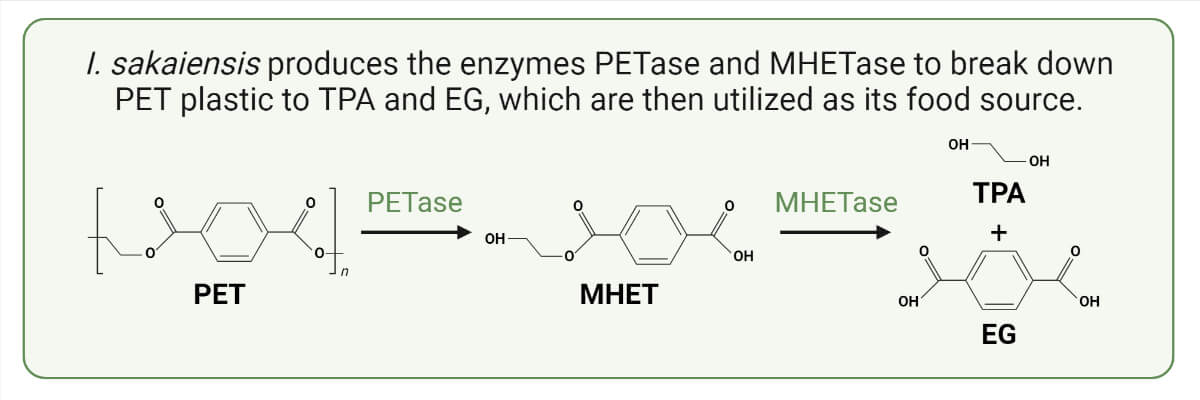
- The bacterial enzyme PET hydrolase, or PETase, breaks down PET into mono (2-hydroxyethyl) terephthalic acid (MHET), a heterodimer made of terephthalic acid (TPA) and ethylene glycol (EG) by the bacterial enzyme PET hydrolase, or PETase.
- I. sakaiensis PETase works by hydrolyzing the ester bonds found in PET.
- I. sakaiensis and many other bacteria can easily absorb and use ethylene glycol among the products but cannot use TPA, so another enzyme called MHET hydrolase is used.
- In other words, the cell utilizes both molecules derived from the PET to produce energy and use them in its metabolic processes.
- In particular, in terms of PET degradation efficiency, the efficiency of this system is among the best at room temperature of known PET hydrolases.
- The assimilation of carbon may eventually result in the mineralization of carbon into carbon dioxide and its release into the atmosphere.
2) MHET hydrolase
- Another PET metabolism-related enzyme that belongs to the tannase family is present in the Ideonella sakaiensis genus.
- The primary PET hydrolysis product of PETase, 2-hydroxyethyl terephthalic acid (MHET), is hydrolyzed by this enzyme into TPA and EG.
- This enzyme was named MHET hydrolase (MHETase) because it is highly active and specific to mono-(2-hydroxyethyl) terephthalate (MHET) but has a negligible hydrolytic effect on bis-(2-hydroxyethyl) TPA (BHET) and several other substrates.
- The crystal structure revealed its composition of a lid domain and a hydrolase domain.
- While the lid domain offers substrate specificity, the hydrolase domain contains amino acid residues necessary for catalysis.
- This enzyme is lipid-anchored on the bacterial cell’s outer membrane and breaks down the resulting MHET into its two monomeric components.
- Here, terephthalic acid (TPA) is imported into the cell through the terephthalic acid transporter protein and is more resistant to degradation.
- Terephthalic acid-1, 2- dioxygenase and 1, 2-dihydroxy-3, 5-cyclohexadiene-1, 4-dicarboxylate dehydrogenase oxidize the aromatic terephthalic acid molecule into a catechol intermediate once it has entered the cell.
- Before the substance enters other metabolic pathways (such as the TCA cycle), PCA 3, 4-dioxygenase cleaves the catechol ring, ensuring that no unsafe or harmful products are left in the ecosystem.
Lab Diagnosis of Ideonella sakaiensis
- The morphological and cultural characteristics of the cells grown on NBRC no. 802 agar plate, including cell shape, colony appearance, and pigmentation, were examined.
- Using transmission electron microscopy, the presence of flagella in cells grown in NBRC no. 802 broth at 30 degrees Celsius was analyzed.
- It was also possible to determine the biochemical properties such as catalase, oxidase, MR–VP, and the hydrolysis of casein, esculin, and starch.
- Also, for further verification of species, Polymerase chain reaction (PCR) is used.
Potent applications of Ideonella sakaiensis
The identification of Ideonella sakaiensis may be significant for the breakdown of PET plastics. Before its discovery, only a small number of bacteria and fungi, such as Fusarium solani, were known PET degraders, and no organisms were definitely known to use PET as a primary carbon and energy source.
- The identification of I. sakaiensis and its ability to assimilate PET through hydrolysis, followed by downstream metabolism of its monomers, may offer a potential remedy for biological recycling of PET, either by using I. sakaiensis directly or its PET-hydrolyzing enzymes through metabolic engineering of other organisms.
- PET’s continued biodegradation could be a key innovative technology for a circular economy and the achievement of Sustainable Development Goals.
- Ideonella sakaiensis and other organisms, along with their hydrolyzing enzymes, would be essential for the development of numerous potential applications.
Future of Ideonella sakaiensis
- The majority of water bottles, polyester clothing, and dinner trays are made of PET, so scientists predict that they will be able to use the bacterial species to facilitate widespread biodegradation.
- If applied commercially, experts believe the microbe could be capable of degrading more than 50 million tons of plastic waste each year all over the world.
- Make plastic into a resource by engineering products.
- Many people also think that it will be easier to find other microbes with similar plastic-degrading traits once more research is done to understand this new classification of bacteria.
- Genetic engineering: Ideonella sakaiensis’s PETase, an enzyme that breaks down PET plastics as well as PEF (polyethylene furanoate) plastics, can be genetically altered and combined with MHETase to break down PET more quickly.