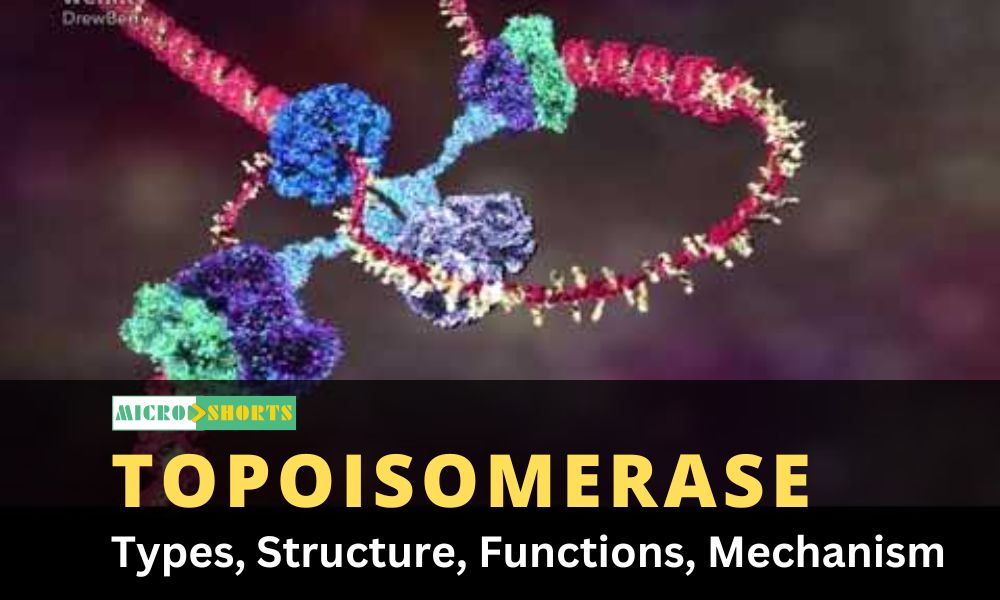
Isomerase enzymes form or assist in forming isomers of any biological components. They assist in the rearrangement process of different biomolecules during the formation or breakage of bonds. Topoisomerase is a type of isomerase enzyme.
What is Topoisomerase?
Topoisomerase is an essential enzyme that aids in the DNA
replication process, segregation of chromosomes, transcription, and also in
recombination.
- It
was first found by J.C. Wang in the 1970s while working on Escherichia
coli. It was the type I topoisomerase.
- As
the name suggests, it helps in changing the DNA topology. It can
increase or decrease the extent of the unwinding of DNA.
- It
can also be called DNA topoisomerase as it only acts on DNA strands.
- It
doesn’t work on RNA.
- It
breaks the phosphodiester bond that is present in the backbone of DNA
strands. The bonds are formed again as the enzyme leaves.
Some Important Terms
Twist (Tw): It is the total count of helical turns of
the strands of DNA.
Writhe (Wr): It is the total count of turns of the
double helix of DNA crossing on itself that indicates the supercoils of DNA.
Linking Number: It is the total number or addition of
twists and writhes in DNA.
Linking No. = Wr+Tw
Topoisomerase Types
There are two types of topoisomerases:
- Type
I Topoisomerase
- Type II Topoisomerase
Type I Topoisomerase
Definition
Type I topoisomerase is a type of topoisomerase that cuts on
a single strand of DNA. It is not an ATP-dependent enzyme(exception: Reverse
Gyrase).
It mainly changes the linking number by plus one.
Note: Odd types of topoisomerases come under type I and
even types under type II.
Type I Topoisomerase Structure
There is the presence of multiple varying domains in the
type IA. It can be from I to IV. Toprim domain is contained in domain I. HTH
(Helix-Turn-Helix) is present in domains III and IV. The tyrosine residues are
present in the HTH of domain III. It appears like a lock with all three domains
present at bottom of the topoisomerase structure.
Type IB contains active site (tyrosine) bind with C-terminal
domain, N-terminal domain, capping, and catalytic lobe.

Figure: Structure of Full-Length Topoisomerase I from Thermotoga
maritima in monoclinic crystal form.
Type I Topoisomerase Types
It is of three basic types:
Type IA topoisomerases
It binds to the 5′ Carbon end of the DNA.
This type of topoisomerase show homology to topoisomerase I
of E. coli.
It is of further three types:
- Topo
IA: It is found in eubacteria.
- Topo
III: It is found in eubacteria and eukaryotes.
- Reverse
Gyrase: It is found in archaebacteria and eubacteria as well. It is the
only type of type I topoisomerase that is ATP-dependent.
(Here topo indicates topoisomerase)
Type IB topoisomerases
It binds to the 3′ Carbon end of the DNA. It forms nick in
one strand. This type of topoisomerase show homology to topoisomerase I of
humans.
Type IC topoisomerases
It contains one type of topoisomerase i.e. topoisomerase V.
It binds to the 3′ Carbon end of the DNA. It is found in archaebacterial. It
shows the controlled mechanism of rotation.
Type I Topoisomerase Mechanism of action
It generally occurs in the following events occurring
together at the same time.
- Cutting
a single strand of DNA: Active site of the topoisomerase contains an
amino acid tyrosine. The disruption of phosphodiester bond and formation
of intermediate with phospho-tyrinosyl linkage favors the breaking of a
DNA strand. The bond formation and cleavage mechanism in detail are the
same as in the case of type II topoisomerase. Tyrosine may attack 3’or
5’carbon end.
- Passing
of strand: After the cleavage, the uncut DNA strand passes
through the break. In this step, the enzyme changes from closed
conformation to open conformation favoring the passing of strand. No ATP
is utilized in this conformational change in the case of type I.
- Religation: The
phosphate linked with tyrosine is again attacked by the OH of the ribose
group of the strand which was separated before and it results in the removal
of intermediate linkage of tyrosine and rejoining of the cleaved strand.
The enzyme returns to its initial stage(closed conformation) and is
recovered for the next cycle.
Type I Topoisomerase Functions
- They
are involved in the removal of supercoils of DNA in biological processes
such as replication and transcription.
- Help
in relaxing DNA.
- They
help in breaking strands during recombination.
- They
are also involved in the condensation of the chromosome.
- During
mitosis, the DNA strands need to be free from interwinding which is done
by topoisomerase I.
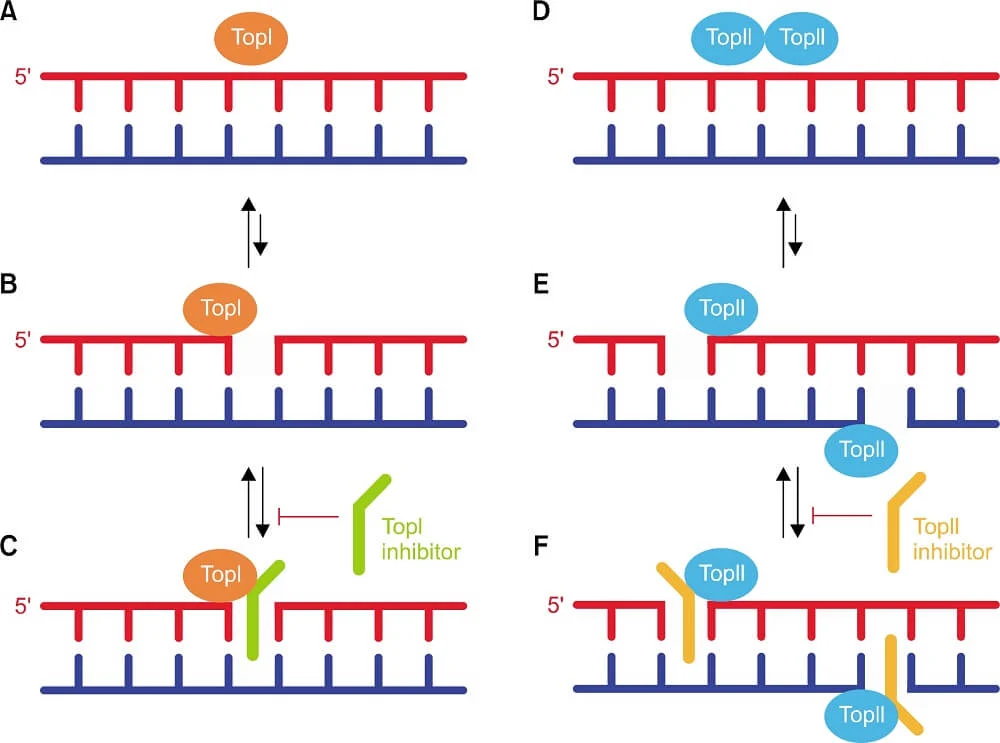
Figure- Topoisomerase I (TopI) and Topoisomerase II (TopII).
Type II Topoisomerase
Definition
Type II topoisomerase is a type of topoisomerase that cuts
on both strands of DNA at once. It is an ATP-dependent enzyme. It changes the
linking number by two.
Type II Topoisomerase Structure
Topoisomerase IIA in eukaryotes consists of two same
monomers (A-A) whereas in prokaryotes they are formed heterotetramers (A2B2).
Topoisomerase IIB is formed of heterotetramers only.
Topoisomerase II consists of four domains which include:
- ATPase
domain at N-terminal
- A
variable C-terminal domain
- Domain
for binding of DNA located centrally
- A
conserved domain of about a hundred amino acids i.e. toprim domain.
Type II Topoisomerase Types
It is of two basic types:
Type IIA topoisomerases
It is found in viruses and all cellular organisms. It is of
three types:
- Topo
II: It is found in eukaryotes.
- Topo
IV: It is found in bacteria. It differs from Gyrase. It is not
involved in DNA wrapping while Gyrase is involved in DNA wrapping and
promoting negative supercoils.
- Gyrase: It
is found in bacteria and some eukaryotes. It introduces negative
supercoiling decreasing the linking number by two.

Type IIB topoisomerases
It includes Topo VI which can be found in archaea and some
plants.

Type II Topoisomerase Mechanism of action
It occurs as follows with ATP hydrolysis.
- Cleaving
of DNA chain: The enzyme contains tyrosine residues. They form
covalent bonds with the DNA strands and break the DNA chain. The lone pair
of electrons of O-atom present in the tyrosine acts as a nucleophile and
attacks on the Phosphorus in phosphate of DNA. It causes the shifting of a
bond from phosphate to one of the O-atom attached to the ribose sugar
forming a hydroxyl group. Hence the covalently bonded tyrosine attached
with phosphorus breaks the phosphate-sugar backbone which cleaves the chain.
This linking is termed 5′-phospho-tyrinosyl protein-DNA linkage.
A duplex is broken by the action of the enzyme on both
strands at once.
- Crossing
of the intact strand through the gap: In this case, another whole
duplex strand passes through the gap over the broken duplex. In this the
conformational change in enzyme requires ATP.
- Religation: It
is done by the attack of 3′-OH of the sugar of separated strand on
phosphate group which has formed an intermediate linkage with tyrosine. It
repels the bond with tyrosine and reforms the broken bond to join again.
It occurs on both strands of duplex together ligating them. The enzymes
regain their conformation and continue the cycle.
Type II Topoisomerase Functions
- It
increases the disentanglement of the chromosome.
- It
does not aid in the supercoiling of DNA but is involved in their
relaxation.
- DNA
gyrase promotes the negative supercoils of DNA.
- One
of the most important functions is that it brings the change of two units
in the linking number of loops in DNA.
Topoisomerase Inhibition
Some chemical components can suppress the action of
topoisomerase and are called topoisomerase inhibitors.
They can interfere with the ligation step of DNA which creates
broken strands in the cell causing the death of the cell by apoptosis.
The topoisomerase inhibition principle is used for the
development of drugs for bacterial infection. It includes antibiotics such as
novobiocin, coumermycin of the class coumarins which interfere in ATP binding
in type II topoisomerases in bacteria leading to its death. It also includes
the quinolone class of antibiotics which prevent the religation of nicked DNA
strands in the last step of the topoisomerase working mechanism.
Chemotherapeutic agents applied for the treatment of cancer
can lead to inhibition of topoisomerase in humans. They can stabilize the
intermediate formed by the linkage of tyrosine of topoisomerase and phosphate
of DNA.

Clinical Significance of Topoisomerase
Many medications work by interfering with type II
topoisomerases in bacteria. These medications include broad-spectrum
antibiotics such as fluoroquinolones. They can make the topoisomerase damage
the DNA.
Cancer cell topoisomerases are targeted by some chemotherapy
medications such as Irinotecan and topotecan for type I and teniposide and
etoposide for type II which are also called topoisomerase inhibitors.
In an autoimmune disorder Scleroderma, the
Anti-topoisomerase antibodies(also called anti-scl-70 antibodies) can be seen
against the topoisomerase I antigen.
Topoisomerase vs Helicase
|
Topoisomerase |
Helicase |
|
It is
involved in the prevention of supercoiling of DNA i.e. decreases tension on
the unwound strands. |
It is
involved in the unwinding of DNA strands. |
|
It works on
DNA only. |
It works acts
on DNA and RNA. |
|
It attacks
the phosphodiester bond in the backbone of DNA. |
It attacks
the Hydrogen bonds between the double strands. |
|
Its two types
are: Type I Topoisomerase Type II Topoisomerase |
Its two types
are: RNA helicase DNA helicase |
Topoisomerase vs Gyrase
|
Topoisomerase |
Gyrase |
|
It includes
different types of enzymes including Gyrase. |
Gyrase is a
type of topoisomerase. |
|
It is a large
class of enzymes. |
It is a type
within the sub-class of Type II topoisomerase. |
|
It is present
in both prokaryotes and eukaryotes. |
It is mostly
present in prokaryotes and only in some eukaryotes. |
|
It maintains
the topology of DNA by the combined functions of different types of enzymes.
It includes both negative and positive supercoiling of DNA. |
Its specific
function is to introduce negative supercoiling in DNA strands rather than to
remove them. |
|
Topoisomerases
may or may not be ATP-dependent. |
Gyrase is an
ATP-dependent type of topoisomerase. |