Introduction
PCR is an enzymatic process in which a specific region of
DNA is replicated over and over again to yield many copies of a particular
sequence.
The most widely used target nucleic acid amplification
method is the polymerase chain reaction (PCR).
This method combines the principles of complementary nucleic
acid hybridization with those of nucleic acid replication applied repeatedly
through numerous cycles.
This method is able to amplify a single copy of a nucleic
acid target, often undetectable by standard hybridization methods, and multiply
to 107 or more copies in a relatively short period.
This thus provides ample target that can be readily detected
by numerous methods.
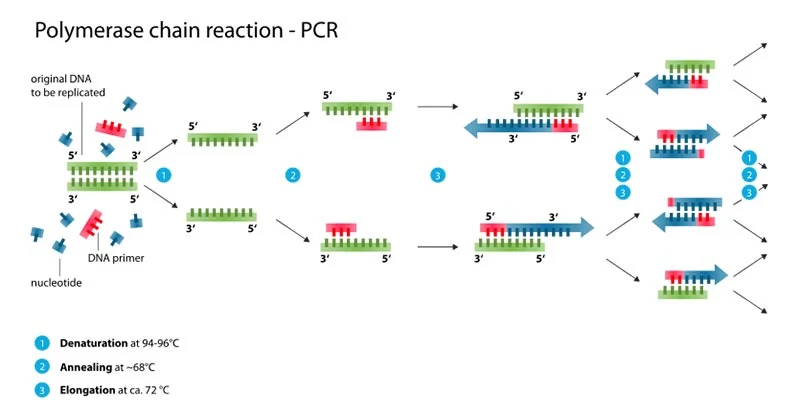
Principle of PCR
The target sequence of nucleic acid is denatured to single
strands, primers specific for each target strand sequence are added, and DNA polymerase
catalyzes the addition of deoxynucleotides to extend and produce new strands
complementary to each of the target sequence strands (cycle 1). In cycle 2,
both double-stranded products of cycle 1 are denatured and subsequently serve
as targets for more primer annealing and extension by DNA polymerase. After 25
to 30 cycles, at least 107 copies of target DNA may be produced
by means of this thermal cycling.
Requirements for PCR
- A
PCR reaction contains the target double-stranded DNA, two primers that
hybridize to flanking sequences on opposing strands of the target, all
four deoxyribonucleoside triphosphates and a DNA polymerase along with
buffer, co-factors of enzyme and water.
- Since
the reaction periodically becomes heated to high temperature, PCR depends
upon using a heat-stable DNA polymerase.
- Many
such heat-stable enzymes from thermophilic bacteria (bacteria that live in
high temperature surroundings) are now available commercially.
- The
first one and the most commonly used is the Taq polymerase from the
thermophilic bacterium Thermus aquaticus.
Procedure of PCR
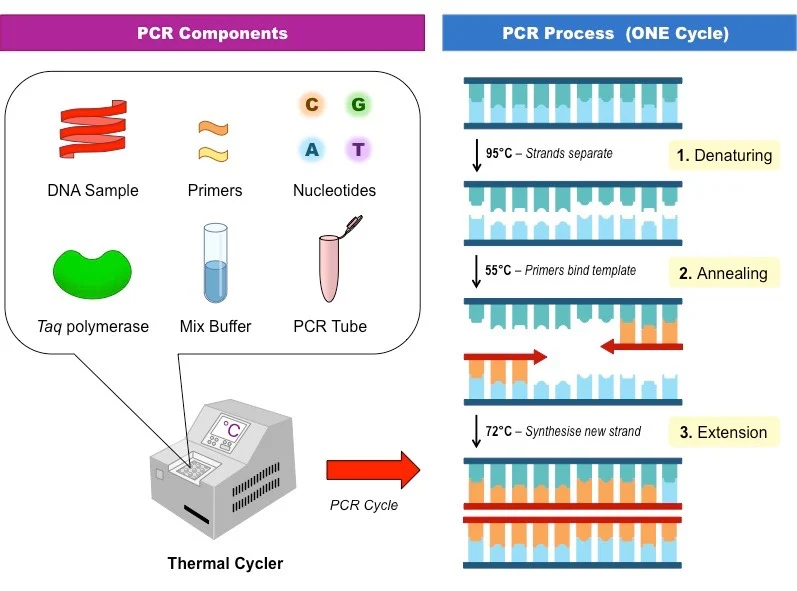
A. Extraction and Denaturation of Target Nucleic Acid
- For
PCR, nucleic acid is first extracted (released) from the organism or a
clinical sample potentially containing the target organism by heat,
chemical, or enzymatic methods.
- Once
extracted, target nucleic acid is added to the reaction mix containing all
the necessary components for PCR (primers, nucleotides, covalent ions,
buffer, and enzyme) and placed into a thermal cycler to undergo
amplification.
B. Steps in Amplification
- Conventional
PCR involves 25 to 50 repetitive cycles, with each cycle comprising three
sequential reactions:
- Denaturation of
target nucleic acid
- Primer
annealing to single-strand target nucleic acid extension of
primer target duplex.
- Extension of
the primer-target duplex.
Denaturation
- The
reaction mixture is heated to 95°C for a short time period (about 15–30
sec) to denature the target DNA into single strands that can act as
templates for DNA synthesis.
Primer annealing
- The
mixture is rapidly cooled to a defined temperature which allows the two
primers to bind to the sequences on each of the two strands flanking the
target DNA.
- Primers
are short, single-stranded sequences of nucleic acid (i.e.,
oligonucleotides usually 20 to 30 nucleotides long) selected to
specifically hybridize (anneal) to a particular nucleic acid target,
essentially functioning like probes.
- This
annealing temperature is calculated carefully to ensure that the primers
bind only to the desired DNA sequences (usually around 55oC).
- One
primer binds to each strand. The two parental strands do not re-anneal
with each other because the primers are in large excess over parental DNA.
Extension
- The
temperature of the mixture is raised to 72°C (usually) and kept at this
temperature for a pre-set period of time to allow DNA polymerase to
elongate each primer by copying the single-stranded templates.
- Annealing
of primers to target sequences provides the necessary template format that
allows the DNA polymerase to add nucleotides to the 3’ terminus (end) of
each primer and extend sequence complementary to the target template
- Taq
polymerase is the enzyme commonly used for primer extension, which occurs
at 72°C. This enzyme is used because of its ability to function
efficiently at elevated temperatures and to withstand the denaturing
temperature of 94°C through several cycles.
- The
ability to allow primer annealing and extension to occur at elevated
temperatures without detriment to the polymerase increases the stringency
of the reaction, thus decreasing the chance for amplification of
non-target nucleic acid (i.e., nonspecific amplification).
The three steps of the PCR cycle are repeated.
- Thus
in the second cycle, the four strands denature, bind primers and are
extended. No other reactants need to be added. The three steps are
repeated for a third cycle and so on for a set of additional cycles.
- By
the third cycle, some of the PCR products represent DNA sequence only
between the two primer sites and the sequence does not extend beyond these
sites.
- As more
and more reaction cycles are carried out, the double-stranded DNA are
synthesized more in number. After 20 cycles, the original DNA has been
amplified a million-fold and this rises to a billion fold (1000) million
after 30 cycles.
C. Product Analysis
- Gel
electrophoresis of the amplified product is commonly employed after
amplification.
- The
amplified DNA is electrophoretically migrated according to their molecular
size by performing agarose gel electrophoresis.
- The amplified DNA forms clear bands which can be visualized under ultra-raviolet (UV) light.
Advantages of PCR
- Amplification
of DNA: PCR allows for the selective amplification of a specific
segment of DNA. This is incredibly useful when working with small or
degraded DNA samples, as it can create enough copies for further analysis.
- High
Sensitivity: PCR can detect a single or a few copies of a target DNA
sequence in a sample. This high sensitivity is crucial in applications
like diagnostics, forensics, and research.
- Speed:
PCR is a relatively fast technique. The entire process can be completed in
a few hours, making it suitable for rapid analysis and diagnostics.
- Versatility:
PCR can be adapted for a wide range of applications, such as genotyping,
DNA sequencing, DNA cloning, and mutation analysis. It can also be used
with different types of DNA, including genomic DNA, cDNA, and RNA.
- Specificity:
PCR primers are designed to be highly specific to the target DNA sequence.
This specificity ensures that only the desired DNA fragment is amplified,
reducing the chance of false-positive results.
- Quantitative
PCR (qPCR): PCR can be modified for quantitative analysis, allowing
researchers to determine the amount of DNA present in a sample. This is
valuable for gene expression studies and quantifying viral or bacterial
load.
- Minimal
Sample Requirements: PCR can work with very small amounts of DNA,
which is particularly advantageous when working with limited or precious
samples.
- Automation:
PCR can be easily automated, reducing the risk of contamination and human
error. High-throughput PCR machines allow for the simultaneous analysis of
many samples.
- Conservation
of DNA: PCR does not consume the entire DNA sample; only the target
region is amplified, leaving the rest of the sample available for future
analyses.
- Low
Cost: PCR is a relatively cost-effective technique, especially when
compared to some alternative methods for DNA analysis.
- Applications
in Medicine: PCR has revolutionized diagnostics in medicine, allowing
for the detection of genetic diseases, pathogens, and cancer markers. It's
also used in prenatal testing and disease monitoring.
- Forensic
Applications: PCR is instrumental in forensic science for DNA
profiling and matching, helping to solve crimes and establish paternity.
- Research
Tool: PCR is a fundamental tool in molecular biology research,
facilitating the study of genes, mutations, and DNA sequences.
- Evolutionary
Studies: PCR is used in the reconstruction of evolutionary
relationships and phylogenetic studies by analyzing DNA sequences from
different species.
- Environmental
Monitoring: PCR can be employed to detect and identify microorganisms
in environmental samples, aiding in pollution assessment and biodiversity
studies.
PCR is a versatile and powerful technique with a wide range of applications in various fields, thanks to its speed, specificity, sensitivity, and ability to work with small sample sizes. These advantages have made it an indispensable tool in molecular biology, medicine, genetics, and many other areas of science and research.
Disadventages of PCR
Sensitivity to Contamination: PCR is highly sensitive, which can be an advantage, but it also makes it prone to contamination. Even trace amounts of foreign DNA, including previously amplified PCR products, can lead to false-positive results. Laboratories must employ stringent contamination control measures.
- Limited Quantitative
Accuracy: PCR is often used for quantitative purposes, but it may not
always provide precise quantitative results. Factors like DNA quality, PCR
efficiency, and the presence of inhibitors can affect the accuracy of
quantification.
- Amplification Bias:
PCR may exhibit amplification bias, meaning it may preferentially amplify
certain DNA sequences over others. This bias can be influenced by factors
like primer design and template secondary structure, potentially leading
to skewed results.
- Limited Size of
Amplifiable DNA Fragments: The size of the DNA fragment that can be
effectively amplified by PCR is limited. Traditional PCR struggles with
very long DNA fragments, and while techniques like long-range PCR exist,
they are not as robust.
- Requirement for DNA
Template: PCR requires a known DNA template to initiate amplification.
This limitation means that it cannot be used for the direct detection of
unknown DNA sequences, making it unsuitable for certain applications like
metagenomics.
- Labor-Intensive:
While technological advancements have streamlined PCR procedures, it can
still be labor-intensive and time-consuming, especially when multiple
cycles are required for amplification.
- Risk of
Cross-Contamination: PCR requires the repeated opening and closing of
reaction tubes, creating opportunities for cross-contamination.
Precautions must be taken to minimize this risk.
- Cost: PCR can be
relatively expensive, particularly when it involves the use of specialized
equipment and reagents. High-quality DNA polymerases, primers, and other
components can also add to the cost.
- Limited Multiplexing:
While multiplex PCR allows the simultaneous amplification of multiple
targets, there is a practical limit to the number of targets that can be
included in a single reaction due to primer interactions and competition
for reagents.
- Evolutionary Analysis
Limitations: PCR is limited in its ability to provide information
about the evolutionary relationships between different DNA sequences. For
this purpose, more advanced techniques like DNA sequencing are often
required.
- Risk of Allelic
Dropout: In some cases, PCR may fail to amplify specific alleles due
to primer-template mismatches, leading to allelic dropout and potentially
misleading results.
- Difficulty with
GC-Rich or Repeat-Containing Regions: PCR may struggle to amplify DNA
regions with high GC content or repetitive sequences due to issues with
primer binding and secondary structure formation.
- Limited to DNA:
PCR is specific to DNA and cannot be directly used for the amplification
of RNA or proteins. Reverse transcription PCR (RT-PCR) is needed to
amplify RNA, and protein analysis requires different techniques.
- Limited Resolution:
In some cases, PCR products of similar sizes may not be easily
distinguishable, limiting its utility in applications where high
resolution is required, such as DNA fingerprinting.
PCR remains an essential tool in molecular biology, genetics, forensics, and diagnostics. Researchers have developed various modifications and alternative techniques to address some of these limitations and expand the range of applications.
Applications of PCR
PCR already has very widespread applications, and new uses
are being devised on a regular basis.
- PCR
can amplify a single DNA molecule from a complex mixture, largely avoiding
the need to use DNA cloning to prepare that molecule. Variants of the
technique can similarly amplify a specific single RNA molecule from a
complex mixture.
- DNA
sequencing has been greatly simplified using PCR, and this application is
now common.
- By
using suitable primers, it is possible to use PCR to create point
mutations, deletions and insertions of target DNA which greatly
facilitates the analysis of gene expression and function.
- PCR
is exquisitely sensitive and can amplify vanishingly small amounts of DNA.
Thus, using appropriate primers, very small amounts of specified bacteria
and viruses can be detected in tissues, making PCR invaluable for medical
diagnosis.
- PCR
is now invaluable for characterizing medically important DNA samples. For
example, in screening for human genetic diseases, it is rapidly replacing
the use of RFLPs.
- Because
of its extreme sensitivity, PCR is now fundamentally important to forensic
medicine. It is even possible to use PCR to amplify the DNA from a single
human hair or a microscopic drop of blood left at the scene of a crime to
allow detailed characterization.