Introduction
There are more than one codon for one amino acid.
This is called degeneracy of genetic code. To explain the possible cause
of degeneracy of codons, in 1966, Francis Crick proposed “the Wobble
hypothesis”.
According to The Wobble Hypothesis, only the first two
bases of the codon have a precise pairing with the bases of the anticodon of
tRNA, while the pairing between the third bases of codon and anticodon may
Wobble (wobble means to sway or move unsteadily).
The phenomenon permits a single tRNA to recognize more than
one codon. Therefore, although there are 61 codons for amino acids, the number
of tRNA is far less (around 40) which is due to wobbling.
The Wobble Hypothesis is a concept in molecular biology that
explains how the genetic code is interpreted during protein synthesis,
specifically in the context of codon-anticodon interactions in the ribosome
during translation. It was first proposed by Francis Crick in 1966.
In the genetic code, a sequence of three nucleotides in a
messenger RNA (mRNA) molecule is known as a codon. Each codon codes for a
specific amino acid, the building blocks of proteins. Transfer RNA (tRNA) molecules
are responsible for carrying the corresponding amino acids to the ribosome
during translation.
The Wobble Hypothesis addresses the phenomenon that there
are more codons (64 possible codons) than there are tRNA molecules carrying
amino acids (around 20 different tRNA molecules). This raises the question of
how the genetic code is read accurately, considering that some tRNA molecules
must be able to recognize and bind to multiple different codons. The Wobble
Hypothesis explains this by introducing a degree of flexibility or
"wobble" in the base-pairing rules between the third position of the
codon (the 5' end) and the first position of the anticodon (the 3' end) of the
tRNA.
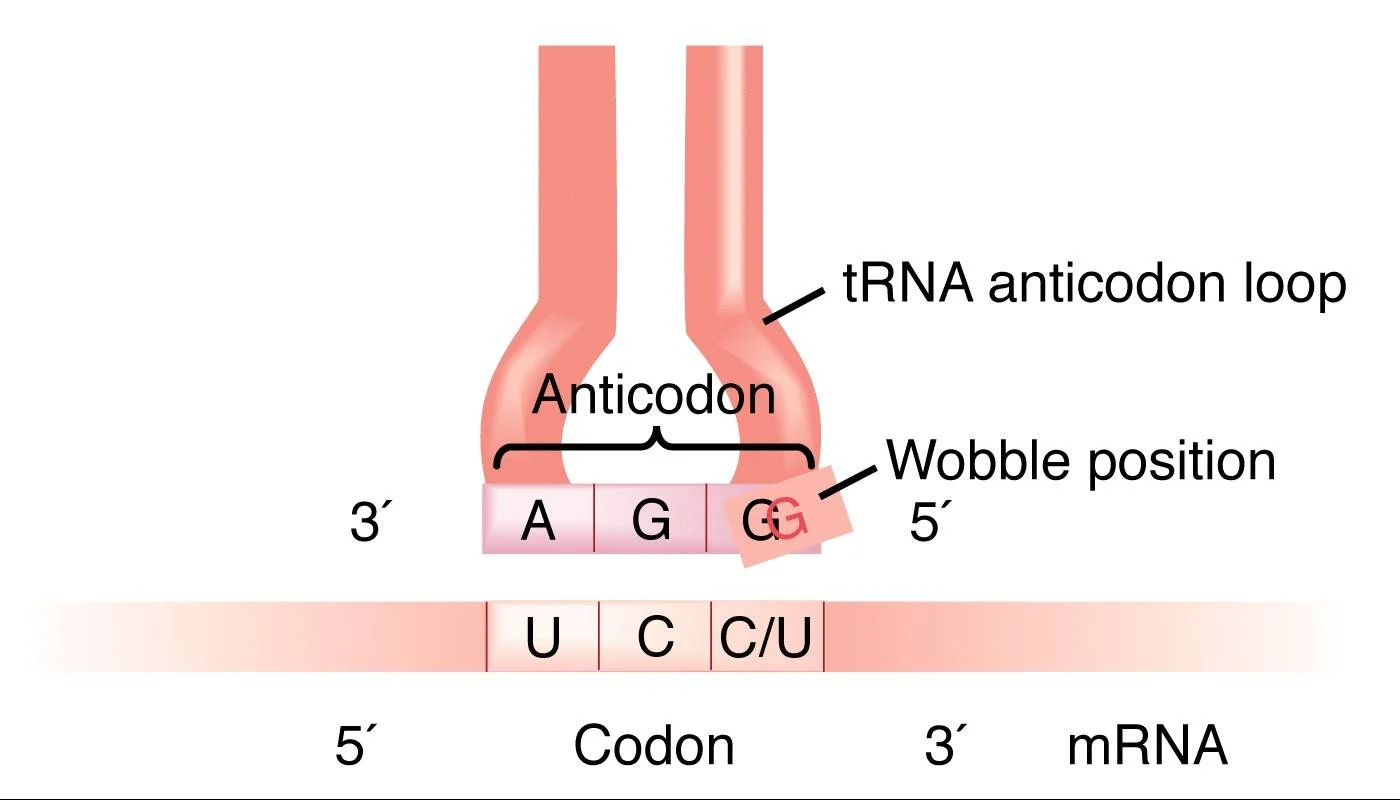
The Wobble Hypothesis Statement
The wobble hypothesis states that the base at 5′ end of
the anticodon is not spatially confined as the other two bases allowing it to
form hydrogen bonds with any of several bases located at the 3′ end of a codon.
This leads to the following conclusions:
- The
first two bases of the codon make normal (canonical) H-bond pairs with the
2nd and 3rd bases of the anticodon.
- At
the remaining position, less stringent rules apply and non-canonical
pairing may occur. The wobble hypothesis thus proposes a more flexible set
of base-pairing rules at the third position of the codon.
- The
relaxed base-pairing requirement, or “wobble,” allows the anticodon of a
single form of tRNA to pair with more than one triplet in mRNA.
- The
rules: first base U can recognize A or G, first base G can recognize U or
C, and first base I can recognize U, C or A.
- Standard
Base Pairing: In the first and second positions of the codon-anticodon
interaction, standard Watson-Crick base pairing occurs. For example,
adenine (A) in the mRNA codon pairs with uracil (U) in the tRNA anticodon
via a complementary base pairing, and guanine (G) in the mRNA codon pairs
with cytosine (C) in the tRNA anticodon.
- Wobble
Base Pairing: In the third position of the codon (5' end) and the first
position of the anticodon (3' end), the base pairing rules are less
stringent. This allows for some degree of flexibility in the pairing. For
example:
- Codons
ending in adenine (A) can pair with tRNAs carrying uracil (U) or adenine
(A) at the first anticodon position.
- Codons
ending in cytosine (C) can pair with tRNAs carrying guanine (G) or uracil
(U) at the first anticodon position.
- Codons
ending in guanine (G) can pair with tRNAs carrying cytosine (C) at the
first anticodon position.
- Codons
ending in uracil (U) can pair with tRNAs carrying adenine (A) or guanine
(G) at the first anticodon position.
Crick’s hypothesis hence predicts that the initial two
ribonucleotides of triplet codes are often more critical than the third member
in attracting the correct tRNA.
Wobble base pairs

- A wobble
base pair is a pairing between
two nucleotides in RNA molecules that does not follow
Watson-Crick base pair rules.
- The
four main wobble base pairs
are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A),
and hypoxanthine-cytosine (I-C).
- In
order to maintain consistency of nucleic acid nomenclature, “I” is used
for hypoxanthine because hypoxanthine is
the nucleobase of inosine.
- Inosine displays
the true qualities of wobble, in that if that is the first nucleotide in
the anticodon then any of three bases in the original codon can be matched
with the tRNA.
Significance of the Wobble Hypothesis
- Our
bodies have a limited amount of tRNAs, and wobble allows for broad
specificity.
- Wobble
base pairs have been shown to facilitate many biological functions, most
clearly proven in the bacterium Escherichia coli.
- The
thermodynamic stability of a wobble base pair is comparable to that of a
Watson-Crick base pair.
- Wobble
base pairs are fundamental in RNA secondary structure and are
critical for the proper translation of the genetic code.
- Wobbling
allows faster dissociation of tRNA from mRNA and also protein synthesis.
- The
existence of wobble minimizes the damage that can be caused by a
misreading of the code; for example, if the Leu codon CUU were misread CUC
or CUA or CUG during transcription of mRNA, the codon would still be
translated as Leu during protein synthesis.
This wobble pairing allows a single tRNA molecule to
recognize multiple codons with similar sequences in the third position,
reducing the requirement for a separate tRNA molecule for each codon. It
provides some redundancy and flexibility in the genetic code, ensuring that the
correct amino acid is incorporated into the growing polypeptide chain during
translation.
The Wobble Hypothesis has since been supported by
experimental evidence and is a fundamental concept in molecular biology,
explaining how the genetic code is efficiently and accurately translated into
proteins despite the redundancy in codon-anticodon interactions.