Introduction
Deoxyribonucleic acid (DNA) is a nucleic acid that is made up of three components: a deoxyribose sugar, a phosphate, and a nitrogenous base. Deoxyribonucleic acid, DNA is the genetic material via which a cell is defined. It is a long molecule containing unique codes that give instructions for the synthesis of all body proteins.
DNA structure
- The
structural model of DNA was initially proposed by James Watson and Francis
Click.
- They
found that DNA is a double-helical structure with two paired DNA strands
with complementary nucleotide sequences.
- The
double-stranded DNA molecule has two spiral nucleic acid chains that are
twisted into a double helix shape. The twisting gives the DNA its
compactness.
- DNA
is made up of millions of nucleotides. Nucleotides are
molecules that are composed of deoxyribose sugar, with a phosphate group
and a nucleobase that is attached to it.
- Each
nucleotide is tightly base paired with a complementary nucleotide on the
opposite strand, i.e Adenine (A) paired with Thymine (T) or Guanine (G)
paired with cytosine (C), and therefore one strand’s sequence acts as a
template for the new strand to be formed during replication.
- Nucleotides
are bound to each other in strands via phosphodiester bonds forming a
sugar-phosphate backbone.
- They
form a bond that is between the third carbon atom on the deoxyribose sugar
made up of one sugar thus it is designated as the 3′ (three prime) and the
fifth carbon atom of another sugar on the next nucleotide as the 5′ (five
prime).
- Any
part of the sequence can be used to create or recognize its adjacent
nucleotide sequence during replication.
- DNA
fits within the nucleus by being closely packed into tight coils known as
chromatins. The chromatins condense to form the chromosomes during cell
division.
- Before
DNA replication, the chromatins loosen up giving the replication machinery
access to the DNA strands.
.webp)
What is DNA Replication?
- This
is a complex process that takes place during cell division, (interphase, S
phase) whereby DNA makes copies (duplicates) before the cell divides
through mitosis and meiosis.
- DNA
replication is a semiconservative process where a
parental strand (template) is used to synthesize a new complementary
daughter strand using several protein elements which include enzymes and
RNA molecules.
- DNA
replication process uses DNA polymerase as the main
enzyme for catalyzing the joining of deoxyribonucleoside 5′-triphosphates
(dNTPs) forming a growing chain of DNA.
- Other
proteins are also involved for initiation of the process and copying of
DNA, along with proofreading capabilities to ensure the replication
process takes place accurately.
- Therefore
DNA replication is a process that produces identical helices of DNA from a
single strand of the DNA molecule.
- DNA
replication is an essential mechanism in enhancing cell growth, repair,
and reproduction of an organism.

Figure: The mechanism of DNA replication. Image
Source: MBInfo © 2018 National University of Singapore.
The mechanism of DNA replication
Summary: DNA replication takes place in three major
steps.
- Opening
of the double-stranded helical structure of DNA and separation of the
strands
- Priming
of the template strands
- Assembly
of the newly formed DNA segments.
- During
the separation of DNA, the two strands uncoil at a specific site known as
the origin. With the involvement of several enzymes and
proteins, they prepare (prime) the strands for duplication.
- At
the end of the process, DNA polymerase enzyme starts to organize the
assembly of the new DNA strands.
- These
are the general steps of DNA replication for all cells but they may vary
specifically, depending on the organism and cell type.
- Enzymes
play a major role in DNA replication because they catalyze several
important stages of the entire process.
- DNA
replication is one of the most essential mechanisms of a cell’s function
and therefore intensive research has been done to understand its
processes.
- The
mechanism of DNA replication is well understood in Escherichia
coli, which is also similar to that in eukaryotic cells.
- In
E.coli, DNA replication is initiated at the oriClocus (oriC), to which
DnaA protein binds while hydrolyzing of ATP takes place.
DNA replication enzymes and Proteins
DNA polymerase
- DNA
polymerases are enzymes used for the synthesis of DNA by adding nucleotide
one by one to the growing DNA chain. The enzyme incorporates complementary
amino acids to the template strand.
- DNA
polymerase is found in both prokaryotic and eukaryotic cells. They both
contain several different DNA polymerases responsible for different
functions in DNA replication and DNA repair mechanisms.
DNA Helicase enzyme
- This
is the enzyme that is involved in unwinding the double-helical structure
of DNA allowing DNA replication to commence.
- It
uses energy that is released during ATP hydrolysis, to break the hydrogen
bond between the DNA bases and separate the strands.
- This
forms two replication forks on each separated strand opening up in
opposite directions.
- At
each replication fork, the parental DNA strand must unwind exposing new
sections of single-stranded templates.
- The
helicase enzyme accurately unwinds the strands while maintaining the
topography on the DNA molecule.
DNA primase enzyme
- This
is a type of RNA polymerase enzyme that is used to synthesize or generate
RNA primers, which are short RNA molecules that act as templates for the
initiation of DNA replication.
DNA ligase enzyme
- This
is the enzyme that joins DNA fragments together by forming phosphodiester
bonds between nucleotides.
Exonuclease
- These
are a group of enzymes that remove nucleotide bases from the end of a DNA
chain.
Topoisomerase
- This
is the enzyme that solves the problem of the topological stress caused
during unwinding.
- They
cut one or both strands of the DNA allowing the strand to move around each
other to release tension before it rejoins the ends.
- And
therefore, the enzyme catalysts the reversible breakage it causes by joining
the broken strands.
- Topoisomerase
is also known as DNA gyrase in E. coli.
Telomerase
- This
is an enzyme found in eukaryotic cells that adds a specific sequence of
DNA to the telomeres of chromosomes after they divide, stabilizing the
chromosomes over time.
Video: DNA replication enzymes and their functions (Shomu’s Biology)
DNA Replication Steps/Stages
Initiation
- This
is the stage where DNA replication is initiated.
- DNA
synthesis is initiated within the template strand at a specific coding
region site known as origins.
- The
origin sites are targeted by the initiator proteins, which
recruit additional proteins that help in the replication process to form a
replication complex around the DNA origin.
- There
are several origin sites on which DNA replication is initiated and they
are all known as replication forks.
- The
formed replication complex contains the DNA helicase enzyme whose function
is to unwind the double helix, exposing the two strands, which act as
templates for replication.
- The
mechanism of DNA helicase enzyme is by hydrolyzing the ATP that is used to
form the bonds between the nucleobases, thus breaking the bond that holds
the two strands.
- Additionally,
during initiation DNA primase enzyme synthesizes small RNA primers that
kick-start the function of DNA polymerase.
- DNA
polymerase enzyme functions by growing the new DNA daughter strand.
Elongation
- This
is the phase where the DNA polymerase grows the new DNA daughter strand by
attaching to the original unzipped template strand and the initiating short
RNA primer.
- The
DNA polymerase is able to synthesize a new strand that matches the
template, by extending the primer via the addition of free nucleotides to
the 3′ end.
- One
of the templates reads in the 3′ to 5′ direction, and therefore, the DNA
polymerase synthesizes the new strand in the 5′ to 3′ direction, which is
known as the leading strand.
- Along
the template strand, DNA primase synthesizes a short RNA primer at the
beginning of the template in the 5′ to 3′ direction, which initiates the
DNA polymerase to continue synthesizing new nucleotides, extending the new
DNA strand.
- The
other template (5′ to 3′) is elongated in an antiparallel direction, by
the addition of short RNA primers which are filled with other joining
fragments, forming the newly formed lagging strand. These
short fragments are known as the Okazaki fragments.
- The
synthesis of the lagging strand is discontinuous since the newly formed
strand is disjointed.
- The
RNA nucleotides from the short RNA primers must be removed and replaced by
DNA nucleotides, which are then joined by the DNA ligase enzyme.
Termination
- After
the synthesis and extension of both the continuous and discontinued
stands, an enzyme knows as exonuclease removes all RNA primers from the
original strands.
- The
primers are replaced with the right nucleotide bases.
- While
removing the primers, another type of exonuclease proofread the new
stands, checking, removing, and replacing any errors formed during
synthesis.
- DNA
ligase enzyme joins the Okazaki fragments to form a single unified strand.
- The
ends of the parent strand consist of a repetition of DNA sequences known
as telomeres which act as protective caps at the ends of chromosomes
preventing the fusion of nearby chromosomes.
- The
telomeres are synthesized by a special type of DNA polymerase enzyme known
as telomerase.
- It
catalyzes the telomere sequences at the end of the DNA.
- On
completion, the parent and complementary strand coil into a double helical
shape, producing two DNA molecules each passing one strand from the parent
molecule and one new strand.
DNA Replication Video Animation (Amoeba Sisters)
Okazaki fragments
- The
two DNA strands run in opposite or antiparallel directions, and therefore
to continuously synthesize the two new strands at the replication fork
requires that one strand is synthesized in the 5’to3′ direction while the
other is synthesized in the opposite direction, 3’to 5′.
- However,
DNA polymerase can only catalyze the polymerization of the dNTPs only in
the 5’to 3’direction.
- This
means that the other opposite new strand is synthesized differently. But
how?
- By
the joining of discontinuous small pieces of DNA that are synthesized
backward from the direction of movements of the replication fork. These
small pieces or fragments of the new DNA strand are known as the Okasaki
Fragments.
- The
Okasaki fragments are then joined by the action of DNA ligase, which forms
an intact new DNA strand known as the lagging strand.
- The
lagging phase is not synthesized by the primer that initiates the
synthesis of the leading strand.
- Instead,
a short fragment of RNA serves as a primer (RNA primer) for the initiation
of replication of the lagging strand.
- RNA
primers are formed during the synthesis of RNA which is initiated de novo,
and an enzyme known as primase synthesizes these short fragments of RNA,
which are 3-10 nucleotides long and complementary to the lagging strand
template at the replication fork.
- The
Okazaki fragments are then synthesized by the extension of the RNA primers
by DNA polymerase.
- However,
the newly synthesized lagging strand is that it contains an RNA-DNA joint,
defining the critical role of RNA in DNA replication.

Figure: Okazaki fragments. Image Source: David O Morgan.
Replication Fork Formation and its function
- The
replication fork is the site of active DNA synthesis, where the DNA helix
unwinds and single strands of the DNA replicates.
- Several
sites of origin represent the replication forks.
- The
replication fork is formed during DNA strand unwinding by the helicase
enzyme which exposes the origin of replication. A short RNA primer is
synthesized by primase and elongation done by DNA polymerase.
- The
replication fork moves in the direction of the new strand synthesis. The
new DNA strands are synthesized in two orientations, i.e 3′ to 5′
direction which is the leading strand, and the 5′ to 3′ orientation which
is the lagging strand.
- The
two sides of the new DNA strand (leading and lagging strand) are
replicated in two opposite directions from the replication fork.
- Therefore
the replication fork is bi-directional.
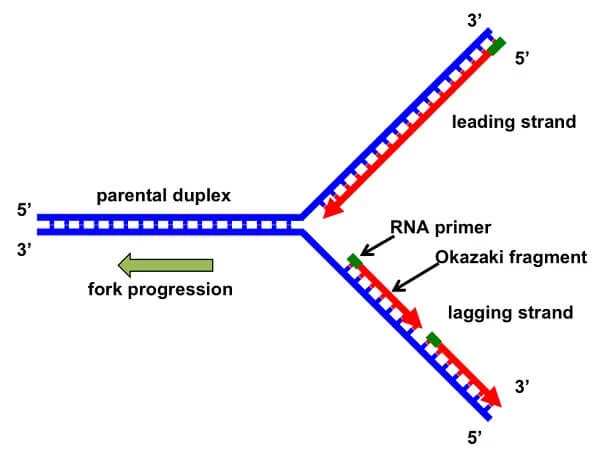
Figure: DNA Replication Fork. Image Source: MDPI (Adam
R. Leman and Eishi Noguchi).
Leading Strand
- The
leading strand is the new DNA strand that is continuously synthesized by
the DNA polymerase enzyme.
- It
is the simplest strand that is synthesized during replication.
- The
synthesis starts after the DNA strand has unzipped and separated. This
generates a short piece of RNA known as a primer, by the DNA
primase enzyme.
- The
primer binds to the 3′ end (start) of the strand, thus initiating the
synthesize of the new strand (leading strand).
- The
synthesis of the leading strand is a continuous process.
The Lagging Strand
- This
is the template strand (5′ to 3′) that is synthesized in a discontinuous
manner by RNA primers.
- During
the synthesis of the leading strand, it exposes small, short strands, or
templates that are then used for the synthesis of the Okasaki fragments.
- The
Okasaki fragments synthesize the lagging strand by the activity of DNA
polymerase which adds the pieces of DNA (the Okasaki fragments) to the
strand between the primers.
- The formation of the lagging strand is a discontinuous process because the newly formed strand (lagging strand) is the fragmentation of short DNA strands.
Why is DNA replication important?
- DNA
replication takes place during cell division and it enables the
multiplication and division of DNA by making two copies of the genome from
a single parent genome.
- And
therefore, its importance is in the creation of new and next copies of DNA
giving rise to two daughter cells from a single parent cell.
- Each
new cell is formed with its own genome.
- This
enhances heredity via reproduction and cell division.
DNA replication stress
During DNA replication, the process and the DNA genome
undergoes various stress arising from the mechanism. these stresses an result
in stalled replication and stalled replication fork formation. Several events
contribute to these stresses, including;
- Unusual
DNA structure
- Mismatched
ribonucleotides
- Tensions
arising from concurrent mechanisms of replication and transcription
- Inadequate
availability of important replication factors
- Fragile
sites on the replicating DNA strand
- Overexpression
or constitutive activation of oncogenes
- Inaccessible
chromatins
Kinase regulatory proteins such as ATM (ATM
serine/threonine kinase) and ATP are proteins that assist in alleviating
replication stress. These proteins get recruited and activated by DNA damages.
Stalled replication forks may collapse if the regulatory
proteins do not stabilize, and if and when this happens, initiation of
repairing mechanisms to reassembling of the replication fork takes place. this
helps to amend damages the damaged ends of DNA.
DNA Replication in Eukaryotes (Differences with
prokaryotes)
DNA replication in prokaryotes and eukaryotes have several
similar features and also differences. This depends on the cell sizes and
genome sizes.
Similarities between Prokaryotic and Eukaryotic DNA Replication
- The
unwinding mechanism of DNA before replication is initiated is the same for
both Prokaryotes and eukaryotes.
- In
both organisms, the DNA polymerase enzyme coordinated the synthesis of new
DNA strands.
- Additionally,
both organisms use the semi-conservative replication pattern, making the
leading and lagging strands in different directions. Okasaki fragments
make the lagging strand.
- Lastly,
both organisms initiate DNA replication using a short RNA primer.
Differences between DNA replication in Eukaryotes and Prokaryotes
|
S.N. |
Eukaryotic DNA Replication |
Prokaryotic DNA replication |
|
1. |
Due to the large size of eukaryotes, they possess 25 times
more DNA |
Due to its small size, they have very minimal/little DNA |
|
2. |
Eukaryotic cells have multiple points of origin and they
use unidirectional replication within the nucleus of the cell. |
Prokaryotic cells have a single point of origin and
replication takes place in two opposite directions at the same time and it
takes place in the cell cytoplasm. |
|
3. |
Eukaryotes have four or more types of polymerases. |
Prokaryotic cells possess one or two types of polymerases. |
|
4. |
Replication of eukaryotic cells is slower taking up to 400
hours. |
Replication in prokaryotic cells is faster, taking up to
40 minutes. |
|
5. |
Eukaryotes have a distinct process for replicating the
telomeres at the ends of their chromosomes. |
Prokaryotes have circular chromosomal DNA therefore they
do not have any ends to synthesize. |
|
6. |
Eukaryotic cells only undergo DNA replication during the
S-phase of the cell cycle. |
Replication in prokaryotes takes place almost
continuously. |