- A set of structural genes (i.e. genes
encoding the proteins to be regulated);
- An operator site, which is a DNA
sequence that regulates transcription of the structural genes.
- A regulator gene which encodes a
protein that recognizes the operator sequence.
Trp Operon
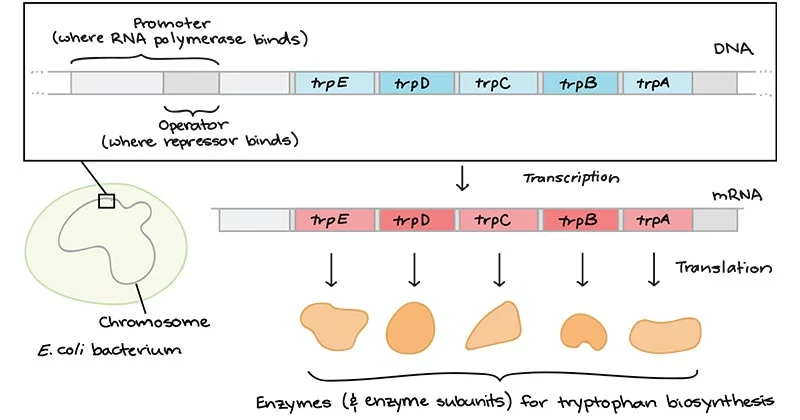
- The
tryptophan operon is the regulation of transcription of the gene responsible
for biosynthesis of tryptophan.
- The
tryptophan (trp) operon contains five structural genes encoding enzymes
for tryptophan biosynthesis with an upstream trp promoter (Ptrp) and trp
operator sequence (Otrp).
- Structural
genes are TrpE, TrpD, TrpC, TrpB and TrpA
- trpE:
It enodes the enzyme Anthranilate synthase I
- trpD:
It encodes the enzyme Anthranilate synthase II
- trpC:
It encodes the enzyme N-5’-Phosphoribosyl anthranilate isomerase and
Indole-3-glycerolphosphate synthase
- trpB:
It encodes the enzyme tryptophan synthase-B sub unit
- trpA:
It encode the enzyme tryptophan synthase-A sub unit
- The
trp operator region partly overlaps the trp promoter.
- The
operon is regulated such that transcription occurs when tryptophan in the
cell is in short supply.
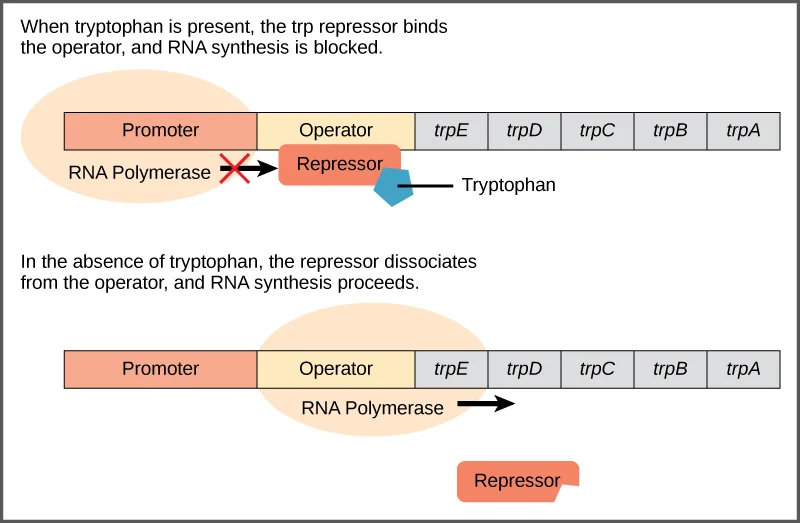
In the Absence of Tryptophan

- In
the absence of tryptophan, a trp repressor protein encoded by a separate
operon, trpR, is synthesized and forms a dimer.
- However,
this is inactive and so is unable to bind to the trp operator and the
structural genes of the trp operon are transcribed.
In the Presence of Tryptophan
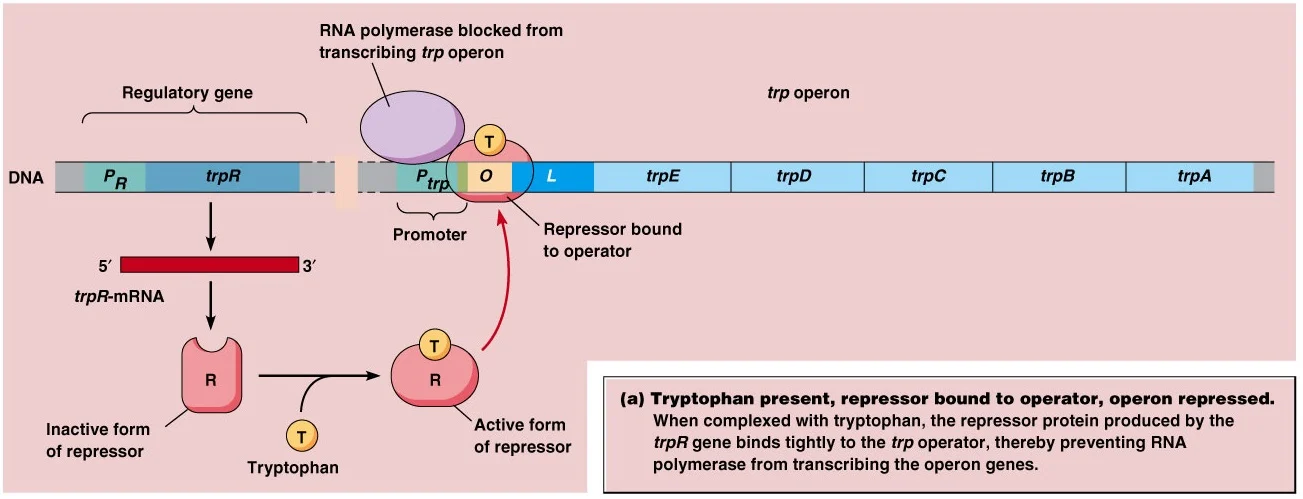
- When
tryptophan is present, the enzymes for tryptophan biosynthesis are not
needed and so expression of these genes is turned off.
- This
is achieved by tryptophan binding to the repressor to activate it so that
it now binds to the operator and stops transcription of the structural
genes.
- Binding
of repressor protein to operator overlaps the promoter, so RNA polymerase
cannot bind to the prometer. Hence transcription is halted.
- In
this role, tryptophan is said to be a co-repressor. This is negative
control, because the bound repressor prevents transcription.
Trp Operon Attenuation
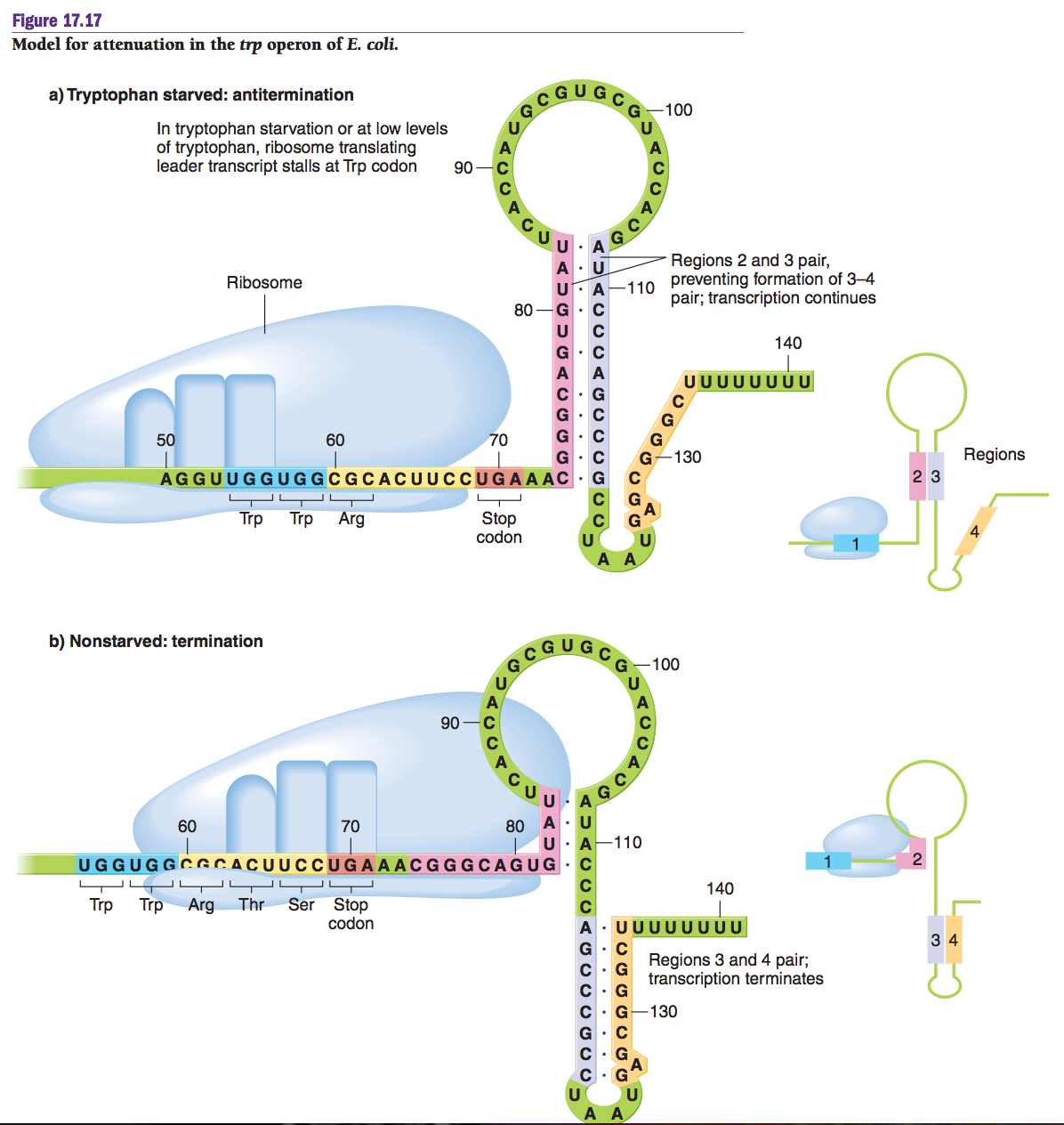
- A
second mechanism, called attenuation, is also used to control expression
of the trp operon.
- The
5′ end of the polycistronic mRNA transcribed from the trp operon has a
leader sequence upstream of the coding region of the trpE structural gene.
- This leader
sequence encodes a 14 amino acid leader peptide containing two
tryptophan residues.
- The
function of the leader sequence is to fine tune expression of the trp
operon based on the availability of tryptophan inside the cell.
The leader sequence contains four regions (numbered 1–4)
that can form a variety of base paired stem-loop (‘hairpin’) secondary structures.
- The
regions are: Region 1, region 2, region 3 and Region 4. Region 3 is
complementary to both region 2 and region 4.
- If
region 3 and region 4 base pair with each other, they form a loop like
structure called attenuator and it function as transcriptional
termination. If pairing occur between region 3 and region 2, then no such
attenuator form so that transcription continues.
Attenuation depends on the fact that, in bacteria, ribosomes
attach to mRNA as it is being synthesized and so translation starts even before
transcription of the whole mRNA is complete.
When Trypophan is abundant

- When
tryptophan is abundant, ribosomes bind to the trp polycistronic mRNA that
is being transcribed and begin to translate the leader sequence.
- Now,
the two trp codons for the leader peptide lie within sequence 1, and the
translational Stop codon lies between sequence 1 and 2.
- During
translation, the ribosomes follow very closely behind the RNA polymerase
and synthesize the leader peptide, with translation stopping eventually
between sequences 1 and 2.
- At
this point, the position of the ribosome prevents sequence 2 from
interacting with sequence 3.
- Instead
sequence 3 base pairs with sequence 4 to form a 3:4 stem loop which acts
as a transcription terminator.
- Therefore,
when tryptophan is present, further transcription of the trp operon is
prevented.
When Trypophan is scarce

- If,
however, tryptophan is in short supply, the ribosome will pause at the two
trp codons contained within sequence 1.
- This
leaves sequence 2 free to base pair with sequence 3 to form a 2:3
structure (also called the anti-terminator), so the 3:4 structure cannot
form and transcription continues to the end of the trp operon.
Hence the availability of tryptophan controls whether
transcription of this operon will stop early (attenuation) or continue to
synthesize a complete polycistronic mRNA.
Regulation of Trp Operon
Overall, for the trp operon, repression via the trp
repressor determines whether transcription will occur or not and attenuation
then fine tunes transcription.
Tryptophan (Trp) Operon
– Many protein-coding genes in bacteria are clustered together in operons which
serve as transcriptional units that are coordinately regulated.
•It was Jacob and Monod in 1961 who proposed the operon model for the
regulation of transcription.
•The operon model proposes
three elements:
•A set of structural genes (i.e. genes encoding the proteins to be regulated);
•An operator site, which is a DNA sequence that regulates transcription of the
structural genes.
•A regulator gene which encodes a protein that recognizes the operator
sequence.
• Operons are thus clusters of structural genes under the control of a single
operator site and regulator gene which ensures that expression of the
structural genes is coordinately controlled.
Trp Operon
• The tryptophan operon is the regulation of transcription of the gene
responsible for biosynthesis of tryptophan.
• The tryptophan (trp) operon contains five structural genes encoding enzymes
for tryptophan biosynthesis with an upstream trp promoter (Ptrp) and trp
operator sequence (Otrp).
• Structural genes are TrpE, TrpD, TrpC, TrpB and TrpA
1. trpE: It enodes the enzyme Anthranilate synthase I
2. trpD: It encodes the enzyme Anthranilate synthase II
3. trpC: It encodes the enzyme N-5’-Phosphoribosyl anthranilate isomerase and
Indole-3-glycerolphosphate synthase
4. trpB: It encodes the enzyme tryptophan synthase-B sub unit
5. trpA: It encode the enzyme tryptophan synthase-A sub unit
• The trp operator region partly overlaps the trp promoter.
• The operon is regulated such that transcription occurs when tryptophan in the
cell is in short supply.
In the Absence of Tryptophan
• In the absence of tryptophan, a trp repressor protein encoded by a separate
operon, trpR, is synthesized and forms a dimer.
• However, this is inactive and so is unable to bind to the trp operator and
the structural genes of the trp operon are transcribed.