Polymerase chain reaction (PCR) is a temperature-dependent nucleic acid amplification technique used to amplify the DNA or RNA in vitro enzymatically.
Developed by Kary Mullis and his associates at mid – 1980s,
it is a very powerful and most important tool in modern biology – molecular
biology and genetics. It combines the principle of nucleic acid hybridization
with the principle of nucleic acid replication. Using this non-culture-based
nucleic acid amplification technique, we can produce billions of copies of a
single segment of DNA or RNA in a very short time.
Since its development, several modifications have been made,
and now there are different types of PCR techniques available for different
purposes. Reverse transcriptase PCR and Quantitative PCR (qPCR) are
the most commonly used PCR types.
Reverse transcriptase polymerase chain reaction, RT-PCR,
is a type of PCR technique that enzymatically amplifies the RNA in vitro.
It is the only type of PCR that can amplify the RNA. It uses
a reverse transcriptase enzyme in addition to the other
basic components of the PCR.
First, the sample RNA is converted to complementary DNA
(cDNA) in reverse transcription, catalyzed by the reverse transcriptase enzyme.
These cDNA molecules are then used as a template for amplification in the PCR
process.
RT-PCR is used to analyze the mRNA or micro RNA and study gene expression.
Objectives of RT-PCR
- To
amplify the specific segment of RNA, resulting in billions of copies of a
single RNA segment.
- To
diagnose certain infections, genes, and study gene expression.
Principle of RT-PCR
RT-PCR combines the reverse transcription
process with the conventional PCR process. The sample RNA is first converted to
double-stranded DNA (complementary DNA) by reverse transcriptase enzyme in the
reverse transcription process. The cDNA can then be thermally broken down into
two single-stranded DNA templates. In these ssDNA templates, primers can anneal
to their complementary sequences based on the nucleic acid hybridization principle.
DNA polymerase then elongates the primer by sequentially adding the nucleotides
to the 3’ end and generates a dsDNA following the principle of DNA replication.
These three processes, denaturation, annealing, and elongation, are repeated in
a cyclic manner regulating the reaction temperature and resulting in millions
of copies of the cDNA.
Requirements (Enzymes) of RT-PCR
1. Nucleic Acid Sample (Sample RNA)
RNA is the sample for RT-PCR, unlike other PCR techniques
using DNA as their sample. Mostly mRNA is used as the sample. The RNA will be
converted into cDNA before amplification.
2. Reverse Transcriptase Enzyme
It is an enzyme that catalyzes the formation of
complementary DNA (cDNA) strands from the RNA strand. It is also called
RNA-dependent DNA polymerase enzyme and is responsible for central-dogma
reverse. It is the major component of RT-PCR as it converts sample RNA into
cDNA for amplification.
3. DNA Polymerase Enzyme
DNA polymerases are enzymes that catalyze the synthesis of
complementary DNA strands by assembling the nucleotides sequentially according
to the template strand. Taq DNA polymerase, the DNA polymerase
enzyme extracted from the bacterium Thermus aquaticus, is the
most widely DNA polymerase as it is thermally stable
and continues its activity after the repeated cycle of heating and cooling.
4. Primers (Oligo (dT) primers, random primers, and sequence-specific primers)
Three different types of primers are used in RT-PCR;
- Random primers
These are the short single-stranded sequences of 6 to 8 nucleotides that bind at the complementary site of RNAs with or without poly(A) for cDNA synthesis using reverse transcriptase.
- Oligo
(dT) Primers
They are oligonucleotides, mostly of 12 – 18 nucleotides,
containing a segment of repeating deoxythymidine (dT) which binds at the polyA
tail of mRNA.
- Sequence-specific
Primers
These are the short single-stranded sequences of nucleotides
that bind to the specific region of interest of the sample RNA. It is mostly
used in one-step RT-PCR.
5. Deoxynucleotide Triphosphates
Deoxynucleotide triphosphates (dNTPs) are artificially
synthesized nucleotides that act as building blocks for synthesizing cDNA and
new cDNA strands during amplification. 4 different dNTPs are used;
deoxyadenosine triphosphate (dATP), deoxyguanosine triphosphate (dGTP),
deoxythymidine triphosphate (dTTP), and Deoxycytidine triphosphate (dCTP).
6. PCR Buffers and Other Chemicals
7. Thermocycler (PCR Machine)
Types of RT-PCR
Based on whether the reverse transcription and the
amplification steps occur either in a single reaction (or tube) or in two
separate reactions (or tubes), RT- PCR can be classified into two types:
1. One-Step RT-PCR
It is a type of RT – PCR where the reverse transcription and
the amplification reactions occur in a single tube. All the required components
are added in a single tube. First, reverse transcription occurs, forming cDNA,
which is then amplified in a PCR process.
Advantages of One–Step RT – PCR over Two–Step RT – PCR
- It
has a simple and easy handling setup.
- It
has higher accuracy and specificity.
- It
has a lesser chance of contamination.
- It
is a cheaper and faster method.
Disadvantages of One-Step RT-PCR over Two-Step RT-PCR
- It
detects fewer templates per reaction mixture due to using multiple
chemicals in a single reaction tube.
- Due
to lower template detection, it requires a larger template for
starting.
- It
does not permit the storage and further analysis of the cDNA formed during
the reaction.
- There
is a higher chance of primer – dimer and non–specific binding.
- The
chance of reaction failure is comparatively high.

One-step vs. Two-step RT-PCR
2. Two-Step RT-PCR
It is another type of RT – PCR where the reverse
transcription and the amplification process occur in two separate tubes. In the
first tube, a reverse transcription reaction takes place, yielding cDNA. These
cDNAs are then transferred to another tube where the PCR mixture is added, and
the cDNAs are amplified.
Advantages of Two-Step RT – PCR over One-Step RT – PCR
- It
allows us to store cDNA formed by reverse transcription.
- It
has higher efficiency, accuracy, and reliability and detects larger
templates per reaction mixture.
- Comparatively,
lower chance of reaction failure, non-specific binding, and primer–dimer
bonding.
Disadvantages of Two–Step RT – PCR over One-Step RT – PCR
- There
is a higher chance of contamination.
- It
is a more complex and tedious process requiring more resources and a
well-trained person.
Steps/Procedure of RT-PCR
The core procedure can be broadly classified into two
phases; reverse transcription and amplification. The procedure also varies on
one-step and two-step RT – PCR. But, the general steps involved in both of the
types are the same and can be summarized into four stages; Preparatory stage,
reverse transcription, amplification, and product analysis stage, viz.:
1. Preparatory Stage
It is the initial stage where RNA extraction is done, and
all the reaction mixture is prepared. First, all materials are arranged, safety
measures are taken, the PCR reaction preparation area is cleaned, all the
reagents are brought to working temperature, and the sample is extracted or
brought from storage.
In the one-step RT-PCR, sample RNA, reverse
transcriptase enzyme, RNase H, primers, DNA polymerase, dNTPs, buffers, and all
other components are added in a specified and pre-calculated amount in a single
reaction tube. The tube is then loaded into a thermocycler for further
processing.
In the two-step, RT-PCR, sample RNA, reverse
transcriptase, RNase H, primers, dNTPs, and other buffers and chemicals for
reverse transcription are loaded in a tube. Then the tube is subjected to a
specified temperature in a thermocycler where cDNAs are
formed.
2. Reverse Transcription
It is the primary step where the RNA is converted into cDNA,
which then undergoes amplification.
All the reaction mixture, including reverse transcriptase,
RNase H, dNTPs mixture, primers, nuclease-free water, reverse transcription
buffer, and other components in one-step RT-PCR and DNA polymerase and other
amplification components in the two-step RT-PCR are added in a tube and
subjected to a temperature of 40 – 50°C for 10 minutes to 30 minutes in
a thermocycler. At this temperature, the primer will bind to the respective
site of the RNA sample, and the reverse transcriptase enzyme will synthesize
cDNA by adding the free dNTPs.
3. Amplification
This step is similar to the amplification process of other
PCR techniques for DNA amplification. In a one-step RT-PCR, the same reaction
mixture is subjected to an amplification process. At the same time, in the
two-step RT-PCR, the cDNA is isolated and placed in another tube where DNA polymerase,
primers, PCR buffer, dNTPs, and other chemicals are added. Then the tube is
placed in a thermocycler for amplification.
The amplification step includes denaturation, annealing, and
elongation occurring cyclically one after another for a certain number of
cycles pre-programmed by the user.
4. Product Analysis Stage
It is the final step where the reaction mixture subjected to
PCR is analyzed to confirm that desired amplification is achieved. The gel
electrophoresis method is mostly used for product analysis. In real-time
RT-PCR, there is no need for this additional step.
Applications of RT-PCR
- Study
Gene Expression
The Traditional Northern Blot technique requires a larger
mRNA sample to analyze and study the gene expression. However, using RT-PCR, we
can amplify the minute mRNA sample and study the sequence of nucleotides, thus
analyzing the gene expression. It is used in studying and identifying
multidrug-resistant genes and their expressions in pathogens.
- Identification
of Unknown Species
RT-PCR is used to identify viruses like HIV, SARS viruses,
dengue viruses, HCV, etc. Besides, other microorganisms and even higher
organisms are identified by studying their rRNA and mRNA.
- Infectious
Disease Diagnosis
Diagnosis of different types of viral infection, bacterial
infection, fungal and parasite infection, cancer cell, and genetic diseases are
done using the RT-PCR technique in clinical laboratories.
- Gene
Insertion and Gene Therapy Study
RT-PCR is used to prepare cDNA from eukaryotic mRNA, which
lacks introns and can be inserted into prokaryotes. RT-PCR is used in
monitoring the result of gene insertion and gene therapy. These procedures are
supposed to show particular gene expression and code for a particular protein,
hence translating specific types of mRNA sequence. This specific mRNA sequence
can be analyzed using RT-PCR.
- Study
Mutation and Cancer Cells
RT-PCR can detect and quantify tissue-specific mutant
alleles. It can also detect any undesired changes in the mRNA sequence and
unique mRNAs, which are produced only by the different types of cancer cells in
our body.
- Tools
of Genetic Engineering and Viral Study
RT-PCR is used in genetic engineering for analyzing modified
DNAs and their transcribed RNAs and amplifying target RNA.
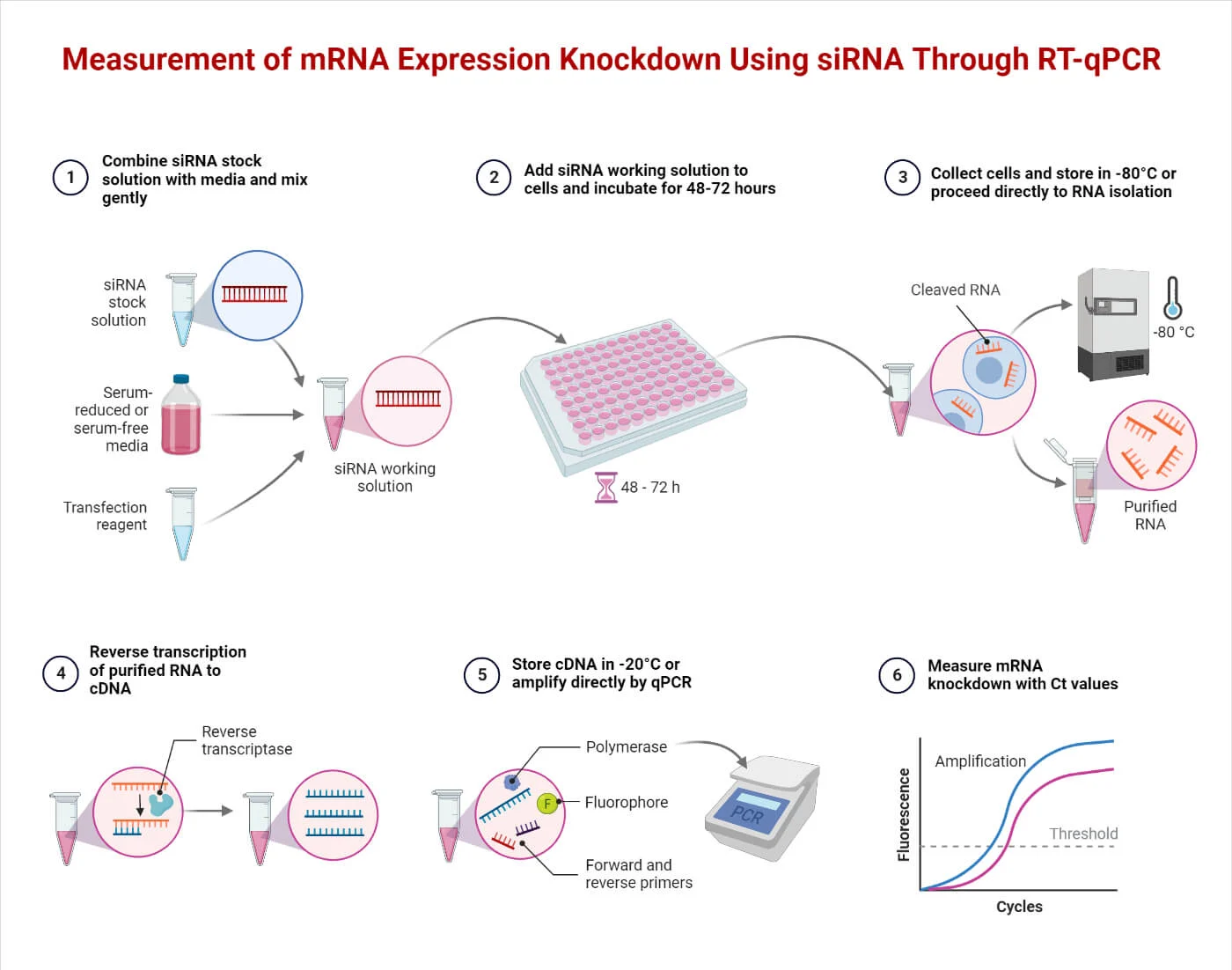
Measurement of mRNA Expression Knockdown Using siRNA Through
RT-qPCR.
Advantages of RT-PCR
- It
is a very rapid method for amplifying RNA and can enzymatically produce
millions of copies of mRNA in a very short time.
- It
is very simple to operate. The process is semi-automatic, operated, and
regulated by a thermocycler without human involvement.
- It
has very high specificity and sensitivity but is economical.
- It
is a very accurate method for the identification of RNA viruses and
infection by them. RNA viruses can be classified up to the level of
strains. It has shortened the time for identifying RNA viruses and viral
infections.
- It
can detect a very minute amount of mRNA (about 5pg) compared to the
traditional Northern Blot technique.
- Mutated
genes and gene expression can be easily and promptly studied. This has
made it possible to diagnose cancer in the early stage, study gene
insertion, and monitor the result of gene therapy.
- It
is both a qualitative and quantitative method; hence can be used to
identify as well as quantify the sample RNA.
Limitations of RT-PCR
- It
can amplify RNAs only, especially mRNAs.
- Prior
information regarding the sequence of the RNA is required for primer
designing.
- It
is a full temperature and enzyme-based system, so a slight change in the
reaction temperature will decrease the efficiency of the enzyme. Hence,
require a strict temperature regulation system.
- Slight
contamination, having a similar primer binding site, can be amplified,
giving a false positive or false negative result.
- The
reaction can be highly influenced by a minute amount of organic or
inorganic contaminant in the reaction mixture.
- The process is very tedious, requiring a complex reaction mixture and a skilled person to operate.