Introduction
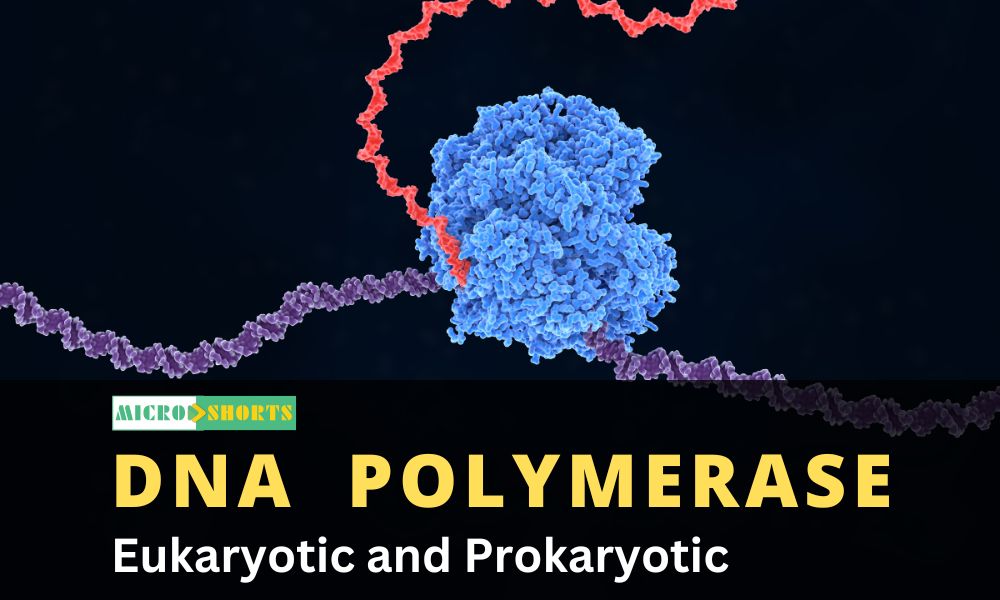
DNA polymerases are a group of enzymes that are used to make copies of DNA templates, essentially used in DNA replication mechanisms. These enzymes make new copies of DNA from existing templates and also function by repairing the synthesized DNA to prevent mutations. DNA polymerase catalyzes the formation of the phosphodiester bond which makes up the backbone of DNA molecules. It uses a magnesium ion in catalytic activity to balance the charge from the phosphate group.
What are DNA polymerases?
- DNA
polymerase was first identified by Arthur Kornberg in lysates of Escherichia
coli, in 1956.
- The
enzyme is found and used in the DNA replication of both prokaryotic and
eukaryotic cells.
- Several
types of DNA polymerase enzymes have been discovered with the first one to
be discovered named DNA polymerase I.
- Each
of these types plays a major role in replication and DNA repair
mechanisms.
- However,
DNA polymerases are not used for initiating the synthesis of new strands,
but in the extension of already existing DNA or RNA strands which are
paired with a template strand.
- DNA
polymerase starts its mechanism after a short RNA fragment is known as a
primer is created and paired with a template DNA strand.
- DNA
polymerase acts by synthesizing the new DNA strand by adding new
nucleotides that match those of the template, extending the 3′ end of the
template chain. Each nucleotide is linked with a phosphodiester bond.
- The
DNA polymerase uses energy from the hydrolysis of the phosphoanhydride
bond that is between the three phosphates (nucleoside triphosphates)
attached to each free base (nucleotides).
- The
addition of a nucleotide to a growing DNA strand forms a phosphodiester
bond between the phosphate of the nucleotide to the growing chain using
the high-energy phosphate bond of hydrolysis, releasing two distal
phosphates known as pyrophosphate.
- DNA
polymerases are very accurate in their mechanism with minimal errors of
less than one error for every 107 nucleotides.
- Some
types of DNA polymerase have the ability to proofread and remove unmatched
bases of nucleotides and correct them.
- They
also correct post-replication mismatches by monitoring and repairing the
errors, by distinguishing mismatches of the new strand from the template
strand sequences.
- The
eukaryotic cell contains five DNA polymerase α, β, γ, δ, and ε. Polymerase
γ is found in the cell mitochondria and it actively replicates the
mitochondrial DNA, while polymerase α, β, δ are found in the cell nucleus
hence are involved in the nuclear DNA replication.
- Polymerase
α and δ are majorly applied and active in diving cells hence involved in
replication while polymerase β is active in both diving and nondividing
cells hence it is involved in the repair of DNA damage.
Structure of DNA polymerase
- The
DNA polymerases generally have a conserved structure, and therefore,
defining its vital role in the cell function which can not be replaced.
- DNA
polymerases are made up of subdomains resembling an open right hand as
palm, fingers, and thumb.
- The
palm contains the catalytic essential amino acids in its active sites.
- The
fingers play a major role in nucleotide recognition and binding.
- The
thumb is for binding of the DNA substrate
- There
is a domain found between the finger and the thumb known as a pocket,
which is made up of two regions i.e the insertion site and the post-insertion
site.
- The
incoming nucleotides bind to the insertion site while the new base pair
bind in the post-insertion site.
- Other
subdomains along with these domains are specific for each family, and each
has essential functions in DNA replication.
- However,
these subdomains are different for each polymerase.
Structure of Family A
- Besides
the already discussed subdomains, Family A polymerase additionally has a
5′ to 3′ exonuclease which is used to remove the RNA primers from the
Okazaki fragments.
- Some
Family A groups also have a 3′ to 5′ exonuclease activity which functions
in proofread the DNA.
Structure of Family B
- They
also possess the basic subdomains with extremely active 3′ to 5;
exonuclease to correct the errors of DNA replication.
Structure of Family X
- These
family groups have the thumb, palm, and finger subdomains which are
structurally part of the N-terminal or on the 31-kDA polymerase fragment.
- The
palm in this family contains three aspartic acid motifs, the fingers have
helices M and N containing amino acid residues.
- The
N-terminal is connected to an 8kDa amino-terminal domain which contains a
5′ deoxyribose phosphate lyase, which is essential in base excision
repair.
Structure of Family Y
- The
N-terminal of this group contains the catalytic core of the palm, fingers,
and thumb.
- They
also have a C-terminal which has a conserved tertiary structure of a
four-stranded beta-sheet supported on one side by two-alpha helices, which
are also known as little finger domain. They play a role in DNA binding
and it is essential for complete polymerase activity.
- However,
this family lacks flexibility, unlike the other families.
DNA polymerase types
Basically, the types of DNA polymerase are also divided
depending on the organism that posses them i.e. eukaryotic and prokaryotic DNA
polymerases. These types of DNA polymerase are classified based on their
characteristics including structural sequences, and functions.
Eukaryotic DNA polymerase types
Polymerase γ
- Polymerase
γ is a Type A polymerase, whose main function is to replicate and repair
mitochondrial DNA.
- It
also functions by proofreading 3′ to 5′ exonuclease activity.
- Mutations
on Poly γ significantly affect the mitochondrial DNA causing autosomal
mitochondrial disorders.
Polymerase α, Polymerase δ, and Polymerase ε
- These
are the type B Polymerase enzymes and they are the main polymerases
applied in DNA replication.
- Pol
α works by binding to the primase enzyme, forming a complex, where they
both play a role in initiating replication. Primase enzyme creates and
places a short RNA primer which allows Pol α to start the replication
process.
- Pol
δ starts the synthesis of the lagging strand from Pol α, while Pol ε is
believed to synthesize the leading strand during replication.
- Studies
indicate that Pol δ replicates both the lagging and leading strand.
- Pol
δ and ε also have a 3′ to 5′ exonuclease activity.
Polymerase β, Polymerase μ, and Polymerase λ
- These
are type 3 or Family X of polymerase enzymes.
- Pol
β has a short-patch base excision repair mechanism where it repairs
alkylated or oxidized bases.
- Pol
λ and Pol μ are important for rejoining DNA double-strand breaks due to
hydrogen peroxide and ionizing radiation, respectively.
Polymerases η, Polymerase ι, and Polymerase κ
- They
are type 4 or family Y polymerases majorly used in repairing of DNA by a
mechanism known as translesion synthesis.
- They
are prone to errors during DNA synthesis.
- Pol
η functions by accurately ensuring the translesion synthesis of DNA
damages that is caused by ultraviolet radiation.
- Pol
κ is still understudies but one of its known functions is to extend or
insert specific bases at certain DNA lesions.
- Translesion
synthesis polymerases are activated by stalled replicative DNA polymerase.
Terminal deoxynucleotidyl transferase (TdT)
- TdT
functions by catalyzing the polymerization of deoxynucleoside triphosphate
to the 3′-hydroxyl group of a preformed polynucleotide chain.
- TdT
is a non-template directed DNA polymerase.
- It
was first detected in the thymus gland.
Prokaryotic DNA polymerase types
DNA Polymerase I
- This
is a type A or Family A polymerase enzyme that was initially isolated
from E. coli and most abundantly found in E.
coli.
- Its
main function is excision repair of DNA strands from the 3′-5′ direction
to the 5′-3 direction, as an exonuclease.
- It
also helps with the maturation of Okazaki fragments, which are short DNA
strands that make up the lagging strand during DNA replication.
- Its
role during replication is the addition of nucleotide at the RNA primer
and it moves along the 5′-3′ direction.
- The
binding site for DNA polymerase I is known as octylglucoside.
DNA Polymerase II
- It
belongs to Type B or Family B of the polymerases.
- Its
major function is the 3′ – 5′ exonuclease activity and to also restart
replication after replication stops due to DNA strand damages.
- The
DNA polymerase II is found in the replication fork, to help in directing
the activities of other polymerases.
DNA Polymerase III
- This
is the primary enzyme that is used in DNA replication, belonging to the
Family C or Type C.
- It
is responsible for the synthesis of new strands by adding nucleotides to
the 3′- OH group of the primer.
- It
has a 3′-5′ exonuclease activity hence it can also proofread the errors
that may arise during DNA strand replication.
DNA Polymerase IV
- It
belongs to the Family Y and it is involved in the non-targeted
mutagenesis.
- Its
activation is based on the stalling activity of the replication fork.
- When
it is activated, it creates a checkpoint, stops replication, and gives
time to properly repair lesions in the new DNA strand.
- It
is also involved in the repair mechanism of translesion synthesis.
- It
does not have nuclease activity, therefore it is prone to errors in DNA
replication.
DNA Polymerase V
- It
belongs to Family Y, with high regulatory activity.
- It
is produced only when DNA is damaged and it requires translesion
synthesis.
- It
also lacks exonuclease functions and hence it can not proofread the synthesis
of DNA replicas making it less efficient.
Taq DNA polymerase
- Taq polymerase is
a thermostable type of DNA polymerase 1 that was initially isolated from a
thermophilic eubacterial known as Thermus aquaticus.
- It
is abbreviated as Taq or Taq pol.
- It
is commonly used in Polymerase Chain Reaction to amplify short strands of
DNA.
- Due
to its thermophilic nature, it is able to withstand denaturation that is
required during PCR, hence it replaced DNA polymerase from E.
coli.
Mechanism of DNA polymerase
The mechanism of DNA polymerase is referred to as a two
metal ion mechanism i.e two metal ions act as active sites for stabilizing the
transmission of replication. The first metal ion acts by activating the
hydroxyl group which attacks the phosphate group of the dNTP and the second
metal ion stabilizes the negative charges and builds on the left oxygen and
chelating phosphate groups.